Open data is a great way of helping give confidence in the reproducibility of research findings. Although we are still a long way from having adequate implementation of data-sharing in psychology journals (see, for example, this commentary by Kathy Rastle, editor of Journal of Memory and Language), things are moving in the right direction, with an increasing number of journals and funders requiring sharing of data and code. But there is a downside, and I've been thinking about it this week, as we've just published a big paper on language lateralisation, where all the code and data are available on Open Science Framework.
One problem is p-hacking. If you put a large and complex dataset in the public domain, anyone can download it and then run multiple unconstrained analyses until they find something, which is then retrospectively fitted to a plausible-sounding hypothesis. The potential to generate non-replicable false positives by such a process is extremely high - far higher than many scientists recognise. I illustrated this with a fictitious example here.
Another problem is self-imposed publication bias: the researcher runs a whole set of analyses to test promising theories, but forgets about them as soon as they turn up a null result. With both of these processes in operation, data sharing becomes a poisoned chalice: instead of increasing scientific progress by encouraging novel analyses of existing data, it just means more unreliable dross is deposited in the literature. So how can we prevent this?
In this Commentary paper, I noted several solutions. One is to require anyone accessing the data to submit a protocol which specifies the hypotheses and the analyses that will be used to test them. In effect this amounts to preregistration of secondary data analysis. This is the method used for some big epidemiological and medical databases. But it is cumbersome and also costly - you need the funding to support additional infrastructure for gatekeeping and registration. For many psychology projects, this is not going to be feasible.
A simpler solution would be to split the data into two halves - those doing secondary data analysis only have access to part A, which allows them to do exploratory analyses, after which they can then see if any findings hold up in part B. Statistical power will be reduced by this approach, but with large datasets it may be high enough to detect effects of interest. I wonder if it would be relatively easy to incorporate this option into Open Science Framework: i.e. someone who commits a preregistration of a secondary data
analysis on the basis of exploratory analysis of half a dataset then receives a code that unlocks the second half of the data (the hold-out sample). A rough outline of how this might work is shown in Figure 1.
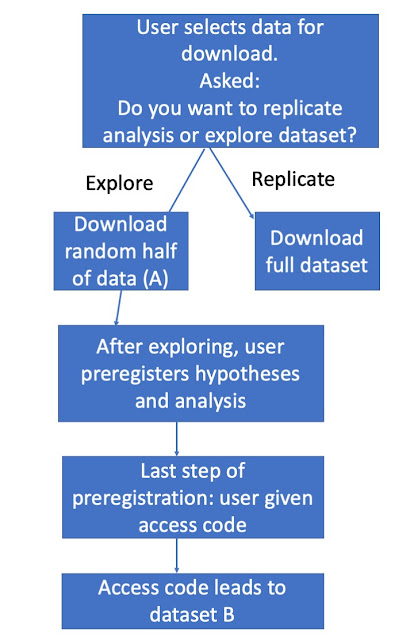 |
| Figure 1: A possible flowchart for secondary data analysis on a platform such as OSF |
An alternative that has been discussed by MacCoun and Perlmutter is blind analysis - "temporarily and judiciously removing data labels and altering data values to fight bias and error". The idea is that you can explore a dataset and run a planned analysis on it, but it won't be possible for the results to affect your analysis, because the data have been changed, so you won't know what is correct. A variant of this approach would be multiple datasets with shuffled data in all but one of them. The shuffling would be similar to what is done in permutation analysis - so there might be ten versions of the dataset deposited, with only one having the original unshuffled data. Those downloading the data would not know whether or not they had the correct version - only after they had decided on an analysis plan, would they be told which dataset it should be run on.
I don't know if these methods would work, but I think they have potential for keeping people honest in secondary data analysis, while minimising bureaucracy and cost. On a platform such as Open Science Framework it is already possible to create a time-stamped preregistration of an analysis plan. I assume that within OSF there is already a log that indicates who has downloaded a dataset. So someone who wanted to do things right and just download one dataset (either a random half, or one of a set of shuffled datasets) would just need to have a mechanism that allowed them to gain access to the full, correct data after they had preregistered an analysis, similar to that outlined above.
These methods are not foolproof. Two researchers could collude - or one researcher could adopt multiple personas - so that they get to see the correct data as person X and then start a new process as person B, when they can preregister an analysis where results are already known. But my sense is that there are many honest researchers who would welcome this approach - precisely because it would keep them honest. Many of us enjoy exploring datasets, but it is all too easy to fool yourself into thinking that you've turned up something exciting when it is really just a fluke arising in the course of excessive data-mining.
Like a lot of my blogposts, this is just a brain dump of an idea that is not fully thought through. I hope by sharing it, I will encourage people to come up with criticisms that I haven't thought of, or alternatives that might work better. Comments on the blog are moderated to prevent spam, but please do not be deterred - I will post any that are on topic.
P.S. 5th July 2022
Florian Naudet drew my attention to this v relevant paper:














One thought: Once more datasets become available, I imagine you can also show that your finding generalizes over several datasets - making it much harder to overfit (still possible o.c.)
ReplyDeleteGreat idea! I think that the "replication" arm should also have a preregistration required (because there are many ways to perform a replication).
ReplyDeleteI came to similar conclusions. For one of my own projects I decided to share synthetic data to help people plan analyses, while retaining full power to run confirmatory analyses. However, there is still a lot of leakage through published analyses and that was a problem in my case. See this post here for some ideas
ReplyDeletehttp://www.the100.ci/2017/09/14/overfitting-vs-open-data/
Since I am suggesting using OSF for this process, I emailed Brian Nosek to see what he thought. He has kindly agreed that I can add his reply here:
ReplyDeleteYes, to your idea of splitting large datasets for reuse. We have done this and, in fact, integrated it with Registered Reports. A small (10%) portion of the dataset is made public for exploration. Authors prepare their Registered Reports. Upon acceptance, we give them the rest of the data for completing the report. In fact, we even hold out an additional 20% of the data for a metascience project in which we are comparing the replicability of the confirmatory analyses in their final RR with any additional exploratory analyses that they included.
The first example of doing this in practice was with the AIID dataset (https://osf.io/pcjwf/). Here's the call for proposals: https://docs.google.com/document/d/1zKqFrMzGsQga7XgBOwWGlmLI4aTfGo4T-xe-Hw-jP5A/edit . I think a total of 15 Registered Reports were completed based on this call.
A team is almost finished preparing another dataset for public dissemination that will do the same thing this year (Ideology 1.0: https://osf.io/2483h/).
These are datasets I created years ago through Project Implicit, and now they are finally getting some real use!
I don't really understand why the best way to prevent p-hacking should be to make it slightly more difficult to p-hack. There are surely other solutions for this; Bayesian analysis isn't going to end all problems, but surely Bayesian model comparing is the way forward when doing hypothesis testing.
ReplyDeleteWouldn't it be better to recognize that when people are doing p-hacking, they are essentially doing exploratory data analysis, and in that case the most likely should be reporting p values in the first place. One way to deal with this is to make reporting p-values more difficult, e.g. by making pre-registration mandatory. Another way would be to actually educate people in how to perform exploratory data analsysis.
On the other hand, having the data and code available is necessary - I often want to look at the data and code, simply because I want to make sure that the authors actually did what they say they did.
I of course meant to write:
Delete"...when people are doing p-hacking, they are essentially doing exploratory data analysis, and in that case they most likely should NOT be reporting p-values in the first place..."